Abstract
Motivation
Radiomics offers a promising non-invasive alternative, leveraging high-dimensional imaging data from routine MR scans to capture the complex biological behavior of gliomas. However, the high dimensionality and intricate spatial structure of glioma radiomics pose significant computational challenges. Our work addresses these challenges by introducing a novel SSM-based approach capable of efficiently processing high-resolution 3D MR images, improving the accuracy of non-invasive tumor subtyping.
Approach
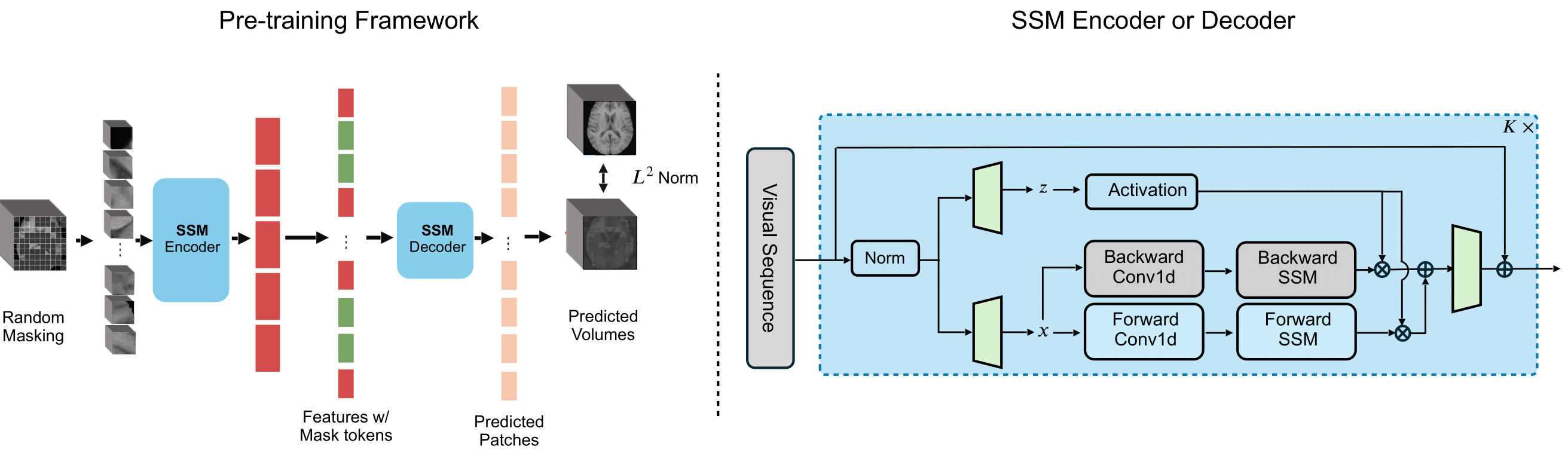 Figure 1: Left: Pre-training a state space model to learn effective representations for 3D multi-contrast MR images. Right: Details of the SSM encoder and decoder. All the vanilla ViT’s attention blocks in MAE are replaced by SSM blocks, while preserving the same pre-training strategy by random masking and reconstruction. The scaled architecture effectively captures global representations for high-resolution data.
Figure 1: Left: Pre-training a state space model to learn effective representations for 3D multi-contrast MR images. Right: Details of the SSM encoder and decoder. All the vanilla ViT’s attention blocks in MAE are replaced by SSM blocks, while preserving the same pre-training strategy by random masking and reconstruction. The scaled architecture effectively captures global representations for high-resolution data.
Interpretable Latent-to-Spatial Map
As illustrated in the below figure, our model’s activations are predominantly concentrated around tumor regions, with the brighter areas indicating regions of interest.
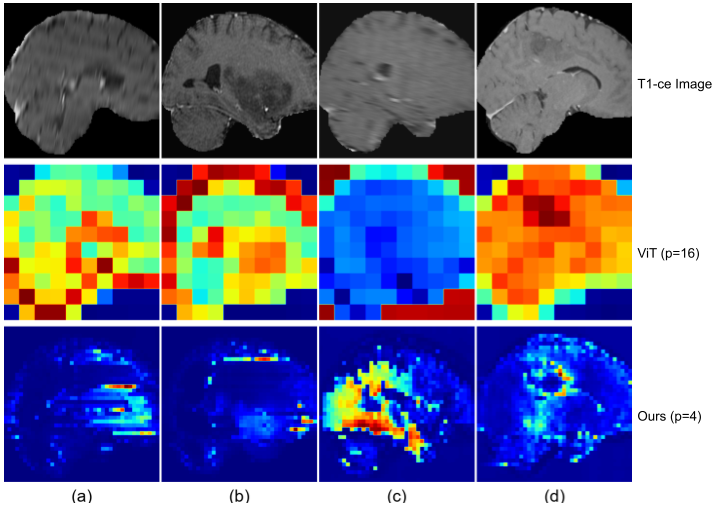
Figure 2: The comparison of saliency maps between the vanilla ViT with a patch size of 16 and our SSM-based model with a patch size of 4. (a) and (b) represent samples with IDH label 0 and 1,respectively; (c) and (b) represent samples with 1p/19q co-deletion label 0 and and 1, respectively.
Linear Scaling Ability
The results below indicate that the linearly scaled model, benefiting from its ability to capture both fine-grained and global features, possesses strong linear scaling capabilities.
5-fold cross-validation results for IDH mutation status classification and 1p/19q co-deletion classification using linearly scaled model across different patch sizes \(p \in\) {4, 16, 32}:
| Patch size | Sequence length | Accuracy (IDH) | F1-score (IDH) | AUC (IDH) | Accuracy (1p/19q) | F1-score (1p/19q) | AUC (1p/19q) |
|---|---|---|---|---|---|---|---|
| 32 | 125 | 0.978 | 0.967 | 0.997 | 0.896 | 0.797 | 0.947 |
| 16 | 1000 | 0.988 | 0.980 | 0.997 | 0.911 | 0.827 | 0.944 |
| 4 | 64000 | 0.998 | 0.997 | 0.999 | 0.911 | 0.832 | 0.958 |
Citation
@misc{hu2024learningbraintumorrepresentation,
title={Learning Brain Tumor Representation in 3D High-Resolution MR Images via Interpretable State Space Models},
author={Qingqiao Hu and Daoan Zhang and Jiebo Luo and Zhenyu Gong and Benedikt Wiestler and Jianguo Zhang and Hongwei Bran Li},
year={2024},
eprint={2409.07746},
archivePrefix={arXiv},
primaryClass={cs.CV},
}